Welcome to DeepGSH
DeepGSH is a web server developed for understanding the S-glutathionylation sites in proteins. Based on deep learning method, the predictor achieved promising performances. Three bright spots of DeepGSH software are summarized below: i) four kinds of protein sequence features are modeled independently and then merged into one model; ii) particle swarm optimizer algorithm is used to optimize the hyperparameters; iii) three fundamental properties of protein sequence are visualized in the result page, including disorder, surface accessibility and secondary structure.
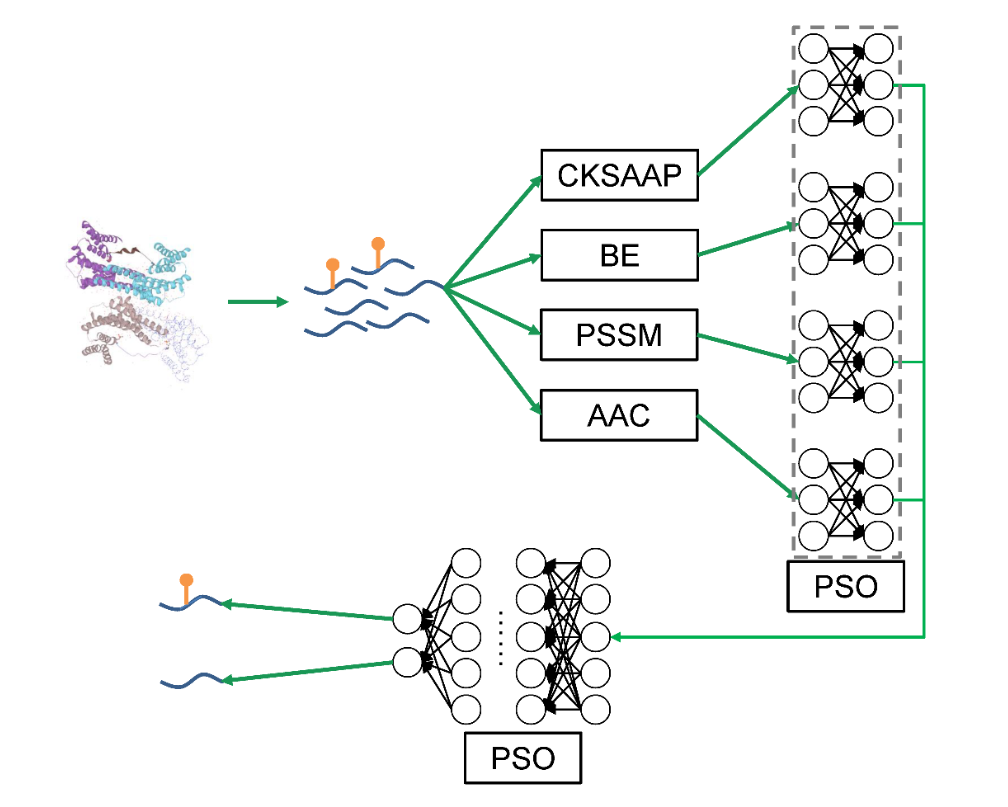
Deep neural network for prediction of S-glutathionylation sites
• Four sequence features of the S-glutathionylation sites are extracted to build models.
• Four models are combined into one full connection layer.
• Particle swarm optimizer (PSO) is applyed to optimize the hyperparameters such as learning rates, activation functions, dropout rates, etc.
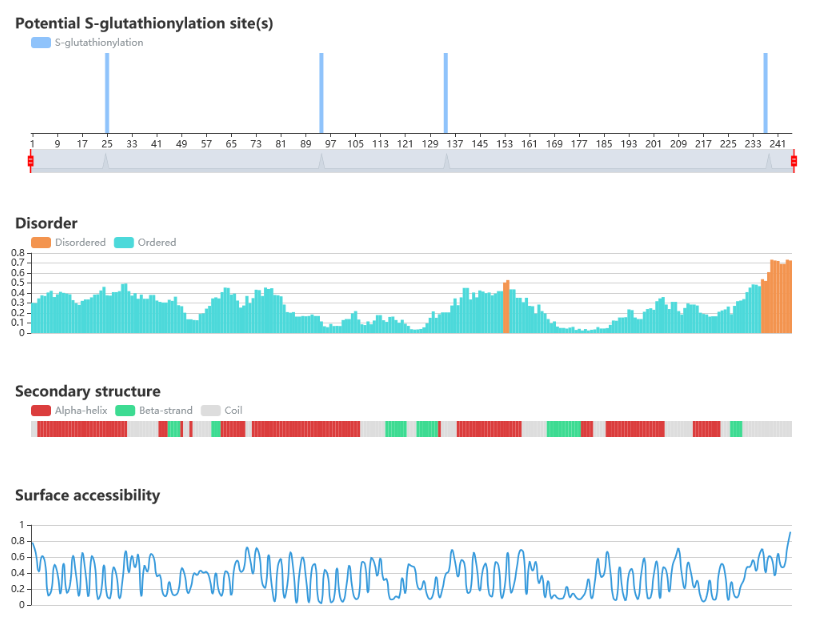
Protein sequence and structure properties
• IUPred is used to predict the disorder region.
• NetSurfP is used to predict the surface accessibility and secondary structure.
• The potential S-glutathionylation sites of the query sequence are indicated.
