Navigation
What is DeepGSH
DeepGSH is a web server based on deep learning to predict S-glutathionylation sites. Given a protein or a set of proteins in FASTA format, it can predict the possible S-glutathionylation sites in proteins. DeepGSH adopted four feature extraction methods to extract the sequence information and used a deep neural network to learn them, in the meanwhile, PSO (particle swarm optimizer) algorithm was used to fine turn the hyperparameters such as learning rate, layer numbers, activation functions and so on. DeepGSH performed better than previous predictors for S-glutathionylation sites.
Usage procedures
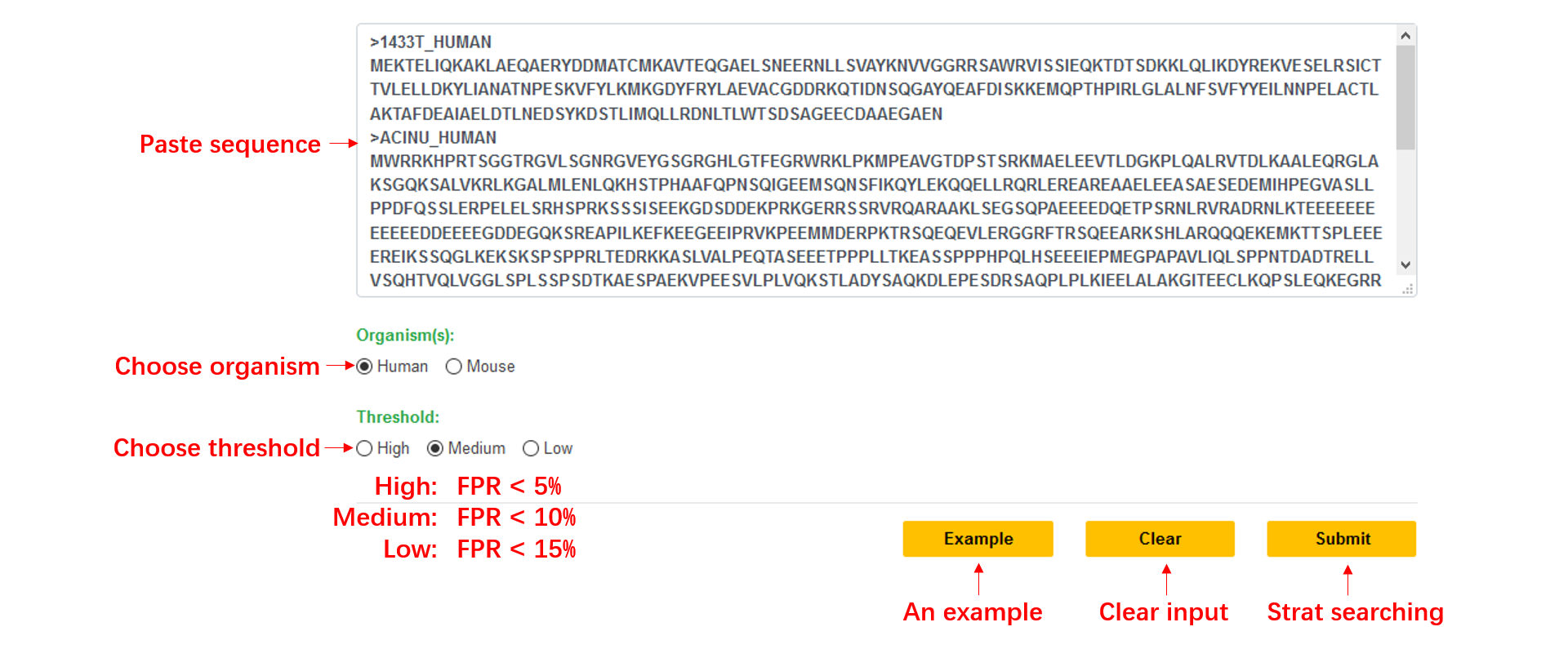
Step1: Paste your FASTA format sequence into the input textarea, or you can click the example button to run the default sequence.
Step2: Choose a kind of organism, the default value is ‘Human’.
Step3: Choose a threshold to get a high confidence result, the default value is ‘Medium’.
Step4: Click Submit.
Output result explanation
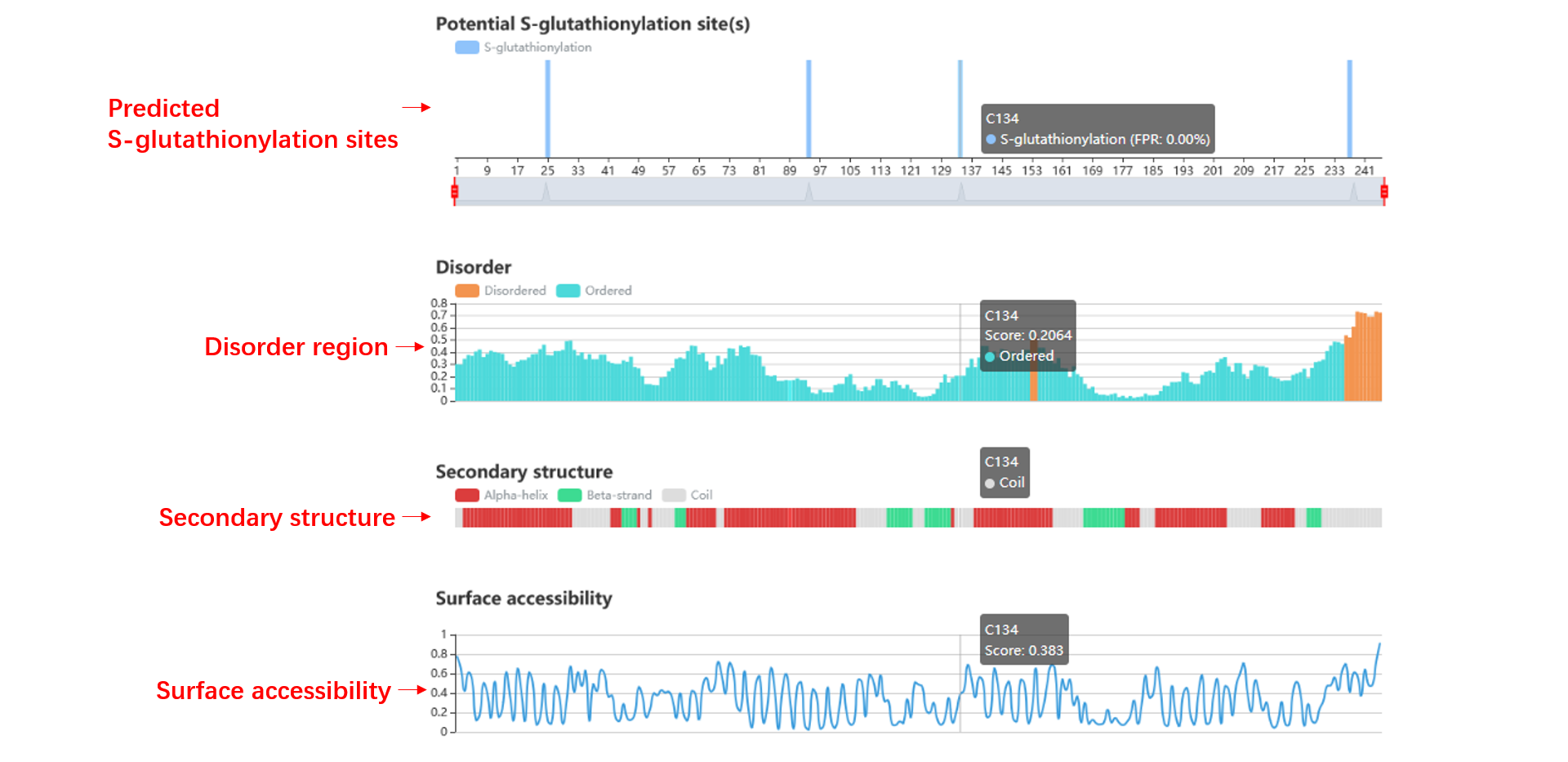
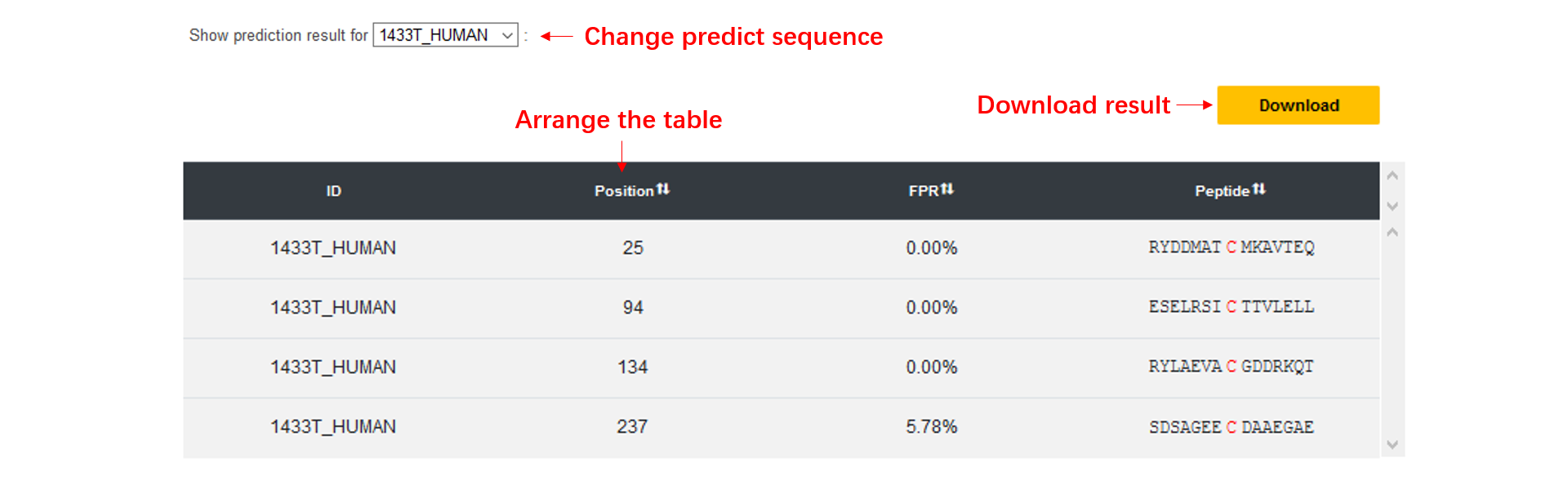
Potential S-glutathionylation site(s)
This table contains four column, including FASTA title ID, position, FPR (false positive rate) and the peptide sequence window.
Secondary structure and surface accessibility
The disorder values are calculated by IUPred. The surface accessibility and secondary structure information are predicted by NetSurfP. By the way, the predicted S-glutathionylation sites are labeled.